Deep Serverless Microscopy
Demo of cellular microscopy using a convolutional neural network and serverless backend.
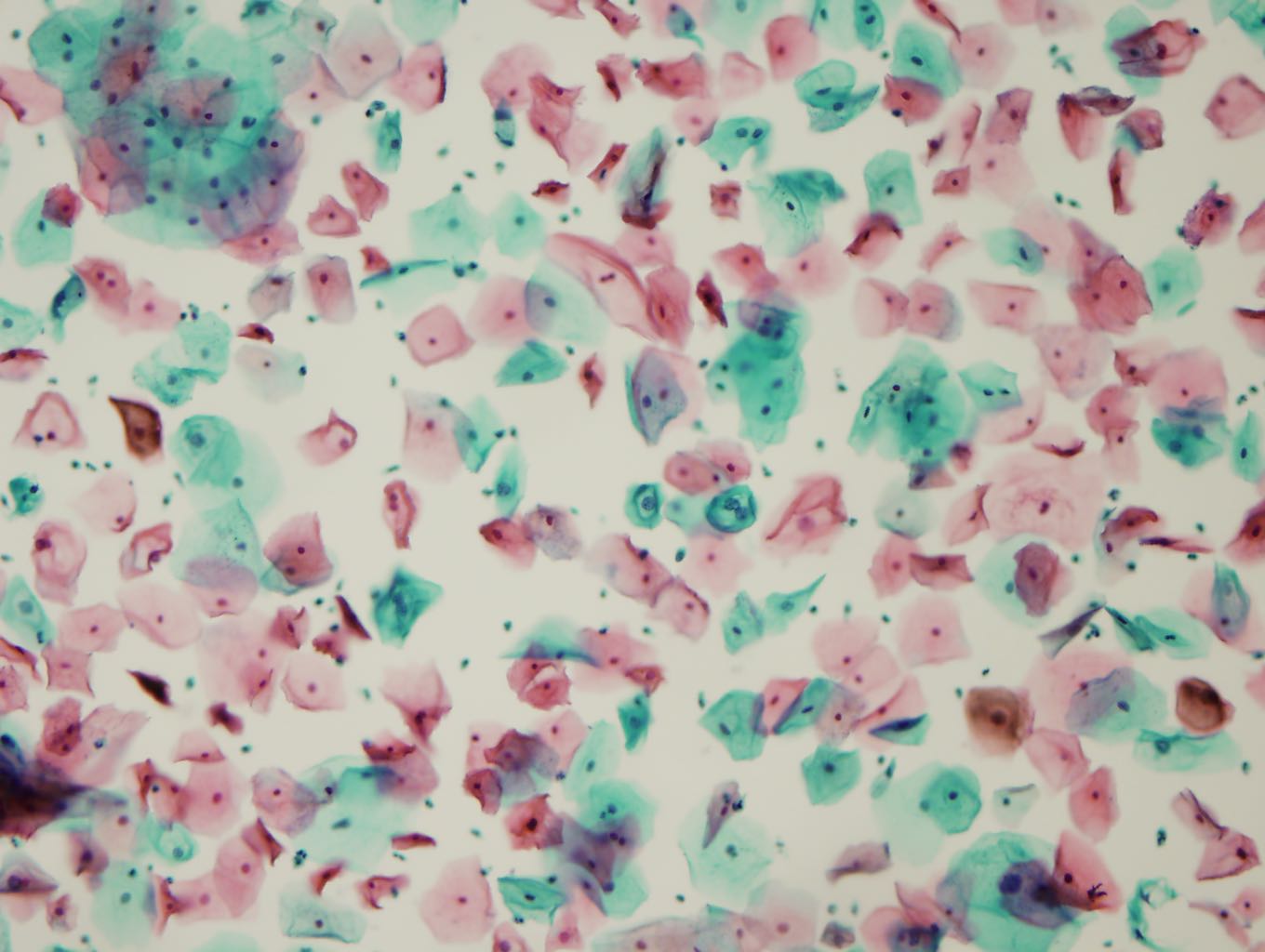
The image on the left is from a precancerous Papanicolaou test. Click on a cell in the image to evaluate it. The first evaluation may take a couple seconds longer as the backend warms up.
How to identify an abnormal cell? Look for rings around the nucleus, multiple nuclei, or grossly enlarged nuclei. Be sure the nuclei are not actually normal parts of two overlapping cells. Hint: See two green cells near the center.
The model is an AlexNet architecture, trained using the Herlev dataset, and implemented in Caffe. Cross-compiled and optimized executable for execution within constrained Lambda environment.
API
Feel free to run the network against your own data. Here is how with curl and python:
curl -X POST -H 'Content-Type: image/jpeg' --data-binary @cell.jpg https://api.hiszpanski.name/cytosight/classify | python -m json.tool